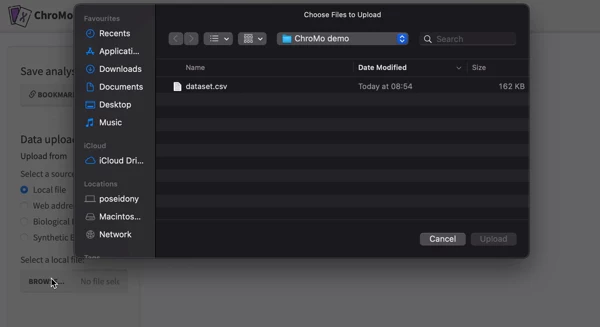
First, data. Everything welcome.
You want to analyze the position and morphology of chromosomes from various cells recorded in vivo. Just upload your measurements from Fiji (or any other tool) in CSV, TSV or XLSX format.
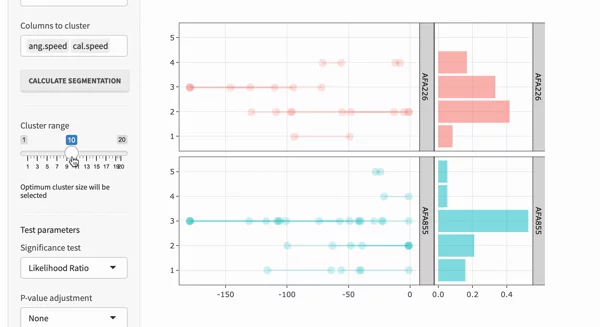
Next, segments. And compare.
Within the time series uploaded, segments are discovered in each of the variables specified. Later, segments are collected and classified into behavioral clusters.
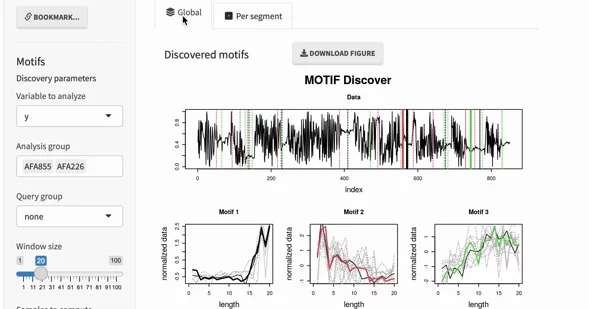
Then, motifs. See for yourself.
Motif (discord) analysis is a tool to consider value sequences and find the most common patterns (and discrepancies) that may appear, at any location.
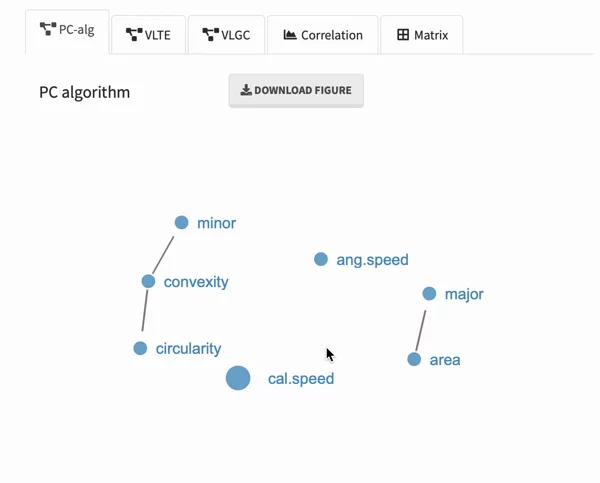
And lastly, causality. All connected.
In ChroMo, we have selected a set of algorithms that allow the study of causality relations across all user-defined variables; that is, whether changes in one variable at a time can explain changes in other variable at a later time.
Available instances
ChroMo S
Small-tier instance
- 256 MB RAM
- 0.25 CPU
- 0 /12 active instances
ChroMo M
Medium-tier instance
- 512 MB RAM
- 0.50 CPU
- 0 /6 active instances
ChroMo L
Large(r)-tier instance
- 1024 MB RAM
- 0.75 CPU
- 0 /2 active instances
Learn more
If you are looking for an in depth explanation of ChroMo and how we used it to explore Chromosome Movements in S. pombe, please read our paper.

If you use ChroMo for your analysis, please cite us:
MDPI and ACS Style
León-Periñán, D.; Fernández-Álvarez, A. ChroMo, an Application for Unsupervised Analysis of Chromosome Movements in Meiosis. Cells 2021, 10, 2013. https://doi.org/10.3390/cells10082013 AMA Style
León-Periñán D, Fernández-Álvarez A. ChroMo, an Application for Unsupervised Analysis of Chromosome Movements in Meiosis. Cells. 2021; 10(8):2013. https://doi.org/10.3390/cells10082013 Chicago/Turabian Style
León-Periñán, Daniel, and Alfonso Fernández-Álvarez. 2021. "ChroMo, an Application for Unsupervised Analysis of Chromosome Movements in Meiosis" Cells 10, no. 8: 2013. https://doi.org/10.3390/cells10082013 BibTeX
@article{LenPerin2021,
doi = {10.3390/cells10082013},
url = {https://doi.org/10.3390/cells10082013},
year = {2021},
month = aug,
publisher = {{MDPI} {AG}},
volume = {10},
number = {8},
pages = {2013},
author = {Daniel Le{\'{o}}n-Peri{\~{n}}{\'{a}}n and Alfonso Fern{\'{a}}ndez-{\'{A}}lvarez},
title = {{ChroMo}, an Application for Unsupervised Analysis of Chromosome Movements in Meiosis},
journal = {Cells}
} ChroMo
ChroMo